Moch R. Adnan 1*; Elly D. I. Wilujeng 1; Mahindra D. N. Aisyah 1; Trisnani Alif 1; Andarula Galushasti 1
1, Department of Agricultural Production, State Polytechnic of Jember, Jember, Indonesia
E-mail:
moch.rosyadi@polije.ac.id
Received: 28/11/2024
Acceptance: 03/02/2025
Available Online: 05/02/2025
Published: 01/07/2025

Manuscript link
http://dx.doi.org/10.30493/DAS.2025.490594
Abstract
The global food security demands an increase in rice production. One of the approaches for improving rice production is through genetic engineering. This study explored the potential of genetic engineering for rice yield improvement by modifying Grassy tiller 1 (OsGT1) gene, a tillering controlling gene. By modulating its expression, rice varieties with an increased (OX) and decreased (Ri) expression of GT1 genes were successfully generated. In this work, a growth characterization experiment was conducted in greenhouse exposed to natural light and additional light exposure for 4 h. The study observed the height of the plant, the length of panicle, number of panicles, number of tillers, and grain weight. The observations indicated that OX lines exhibited significant suppression in tiller number, which might potentially allocate maximum photosynthate to grain production. In contrast, the Ri lines exhibited an increased number of tillers. Moreover, the weight of 1000 grain was significantly lower in Ri plants compared to OX and wild type plants. Although the current findings indicate the potential of GT1 modification to enhance rice productivity, additional research is required to convert these results into a comprehensive increase in rice yield. Comprehending the intricate linkages between GT1 and its vital role in influencing agronomic phenotypes and performances can illustrate how the manipulation of tiller-controlling genes substantially impacts rice productivity.
Keywords: Rice, Tiller number, OsGT1, Greenhouse
Introduction
Rice is a staple crop that fulfills a great portion of the global food demand. Rice cultivation is perpetually subjected to numerous environmental problems that can adversely affect productivity and quality. Consequently, the creation of novel rice cultivars that withstand environmental adversities and yield abundantly is crucial for sustainable agriculture. Yields in rice are influenced morphologically by several agronomic characteristics, including the number of grains per panicle, grain size, grain weight, plant architecture, and tiller count [1][2]. Consequently, employing biotechnology to modify rice architecture, including tillering phenotypes, may represent a promising advancement.
The tiller is the basal stem “branch” of rice that emerges from the axillary bud during the vegetative stage. The juvenile axillary bud subsequently matures into a tiller and primarily ceases growth throughout the generative stages. [3][4]. The alteration of tiller quantity has been associated with an enhancement in potential panicle count and a reduction in nonproductive tiller quantity [5,6]. Consequently, employing genetic engineering to modify tiller quantities may positively affect yield [7].
In rice, Grassy Tiller 1 (GT1), also identified as Homeobox Leucine Zipper12 (HOX12), is a gene that encodes a transcription factor involved in multiple growth and developmental processes. This gene is part of the homeobox transcription factor family, which primarily governs plant architecture by modulating tiller bud outgrowth [8-10]. Previous research has revealed that OsGT1 (Grassy tiller 1) gene has a role in regulating tillering phenotype in rice [11]. A disparity in tillering phenotypes was observed between rice that overexpresses and suppresses the GT1 gene. Transgenic plants with higher expression of GT1 harbored fewer tiller phenotypes, while lower expression of GT1 rendered plants with prolific growth of tillers compared to wild-type. Given that tillers predominantly develop during the vegetative stage, external and internal stimuli influencing the transition from vegetative into generative stage would likely have an impact on the tiller development [12]. Markedly, the alteration of the photoperiod from a subtropic-cultivated variety into tropical setting can significantly affect the tiller formation.
This study used GT1 overexpression (OX) and RNAi (Ri) lines grown in standard greenhouse cultivation system. This research seeks to deliver an initial investigation report on the growth characteristics of OsGT1 mutants, which were generated by previous research using Japonica rice, while cultivated under greenhouse conditions. A supplementary bioinformatic study was also conducted to illustrate GT1 amino acid alignment and phylogeny relations between prominent Poaceae crops, as well as illustrating the used transgenic vector constructs in the assessed transgenic plants.
Material and Methods
Plant materials
Plant materials were obtained from a previous study [11], which encompassed three varieties: wild-type non-transgenic plants of the Japanese rice cultivar ‘Oryza sativa cv. Nipponbare’ (NB), transgenic GT1 overexpression lines (OX), and transgenic gt1-RNAi (Ri). Plants were self-propagated in greenhouse facility located in Politeknik Negeri Jember to produce seeds for the entire experiment.
Bioinformatic assessment
Amino acid sequence alignment and phylogenetic analysis of GT1
Amino acid sequences of GT1 gene in rice, maize, sorghum, Brachypodium distachyon, barley,and wheat were acquired from Rice Annotation Project Database [13], Maize Gramene [14], Sorghumbase [15], EnsemblPlants [16], and Phytozome13 [17], respectively. The amino acid sequences were then aligned and the phylogenetic tree was constructed using the Universal Protein Resource (UniProt) tools [18].
Transgenic vector constructs
The illustration of vectors used for generating transgenic plants based on [11] were visualized using Snapgene software version 2.3.2. Vectors illustrations were generated using pCAMBIA1381 backbone database downloaded from Snapgene Plasmid Collection [19]. Subsequently the transgenic lines constructs were created by placing rice ubiquitin promoter upstream of intact GT1 gene (GT1-OX) and placing rice intron flanked by the sense and antisense strand of 355 bp of C-terminal end of coding sequences of GT1 gene (GT1-Ri).
Cultivation and agronomical phenotyping of transgenic plants
The plant materials were planted in a 5×12 m greenhouse in Politeknik Negeri Jember, Jember, East Java, Indonesia. Before planting, all rice seeds were immersed in ample water for 3-5 days with daily replacement. Subsequently, the seeds were transferred into soil in tray. After 6 weeks, rice seedlings were transferred into pots filled with a mixture of soil and manure 3:1 (approximately 4 Kg) media with dimension of 24×18 cm. The plants were exposed to natural light and an additional artificial light exposure period of 4 h. To maintain a proper plant growth, balanced NPK and Urea fertilizers were implemented to individual plants. A 0.59 g NPK and 0.2 g Urea were administered in two portions: 50% NPK + 70% Urea at 15 days after planting (DAP) and the remaining 50% NPK + 30% Urea at 30 DAP.
To prevent the diseases outbreaks, preventive pesticides were applied regularly in 7-10 days intervals. The utilized pesticides including insecticide (Marshall, Regent 50, BPMC), fungicide (Fujiwan 400, Heksa, Moltovin), bactericide (Bactocyn) were applied according to manufacturers’ recommendation.
Agronomical properties were measured using 8 pots replications for each variety (type). All observations were made at the maturity phase of the plants (160 DAP). Plant heights were measured from the base of stem to the top panicle. Panicle lengths were measured from the base to the tip of top primary branch of panicles. Number of panicles and tillers were manually observed and counted per plants. Number of productive tillers were obtained from the sum of tiller-bearing panicles per total panicles. Moreover, thousand grains weights were assessed
Statistical analysis
The statistical analyses of agronomical observations were carried out in GraphPad Prism 8.01 software using one-way analysis of variance (One-way ANOVA) followed by Tukey’s Multiple Comparison test at p<0.05.
Results and Discussion
OsGT1 gene and its relationship with its homolog genes
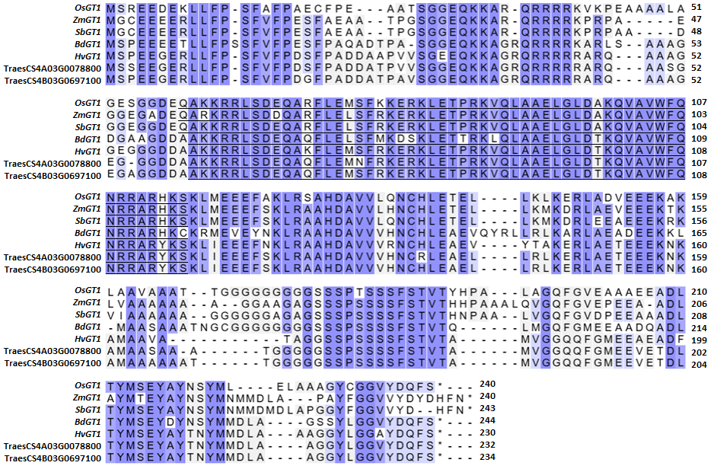
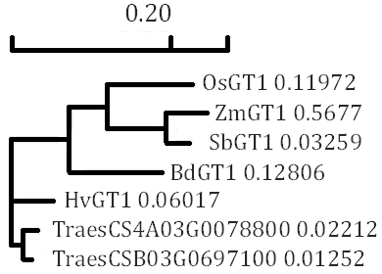
Grassy tiller 1 is a gene initially identified through research on the tillering trait of contemporary maize (Zea mays) and its wild ancestor (Teosinte) [19]. Initial studies on maize indicated that ZmGT1 contributes to the inhibition of tiller bud development in contemporary maize [20]. The low expression of ZmGT1 in its parent, Teosinte, was elucidated to correlate with many tillers and tassels, accompanied by reduced silk and kernel size. Conversely, the elevated expression of GT1 resulted in a reduced number of tillers and tassels, while increasing kernel size; however, evolutionary studies indicate that GT1 is not the only transcription factor selected during maize domestication [21]. A comparable tendency of tillering was observed in rice GT1 (OsGT1). Consequently, examining its origin is essential for ascertaining its role in other crops, including rice [9][11]. The amino acid sequence alignment (Fig. 1) indicated that the OsGT1 had similarities of 81.50% with maize, 83.59% with sorghum, 78.91% with purple false brome, 77.95% with barley, in addition to 79.69% and 77.95% with wheat. Amino acid alignment corroborated previous studies indicating that maize GT1 had a close connection with GT1 of other crops, including rice, with bootstrap values between 95% and 100% [22]. Phylogeny tree of GT1 orthologue gene (Fig. 2) showed that GT1 in rice, maize, and sorghum are clustered inside a single clade. Concurrently, purple false brome, barley, and wheat were collected within a separate lineage. A greater percentage of amino acid similarity indicates an increased likelihood of exhibiting analogous biological activities and functions. OsGT1 is classified as a class I Homeodomain Leucine Zipper (HD-ZIP) that performs various functions in plant growth and development [23]. Class I HD-ZIP transcription factors generally comprise a homeodomain (HD) box serving as the DNA binding domain that enhances gene transcription, and a Leucine Zipper domain (LZ or ZIP) that enables protein dimerization [24]. OsGT1, along with other Hox family proteins in rice, has been previously identified as contributing to panicle exertion, shorter panicle length, and a dwarf plant phenotype [9][25].
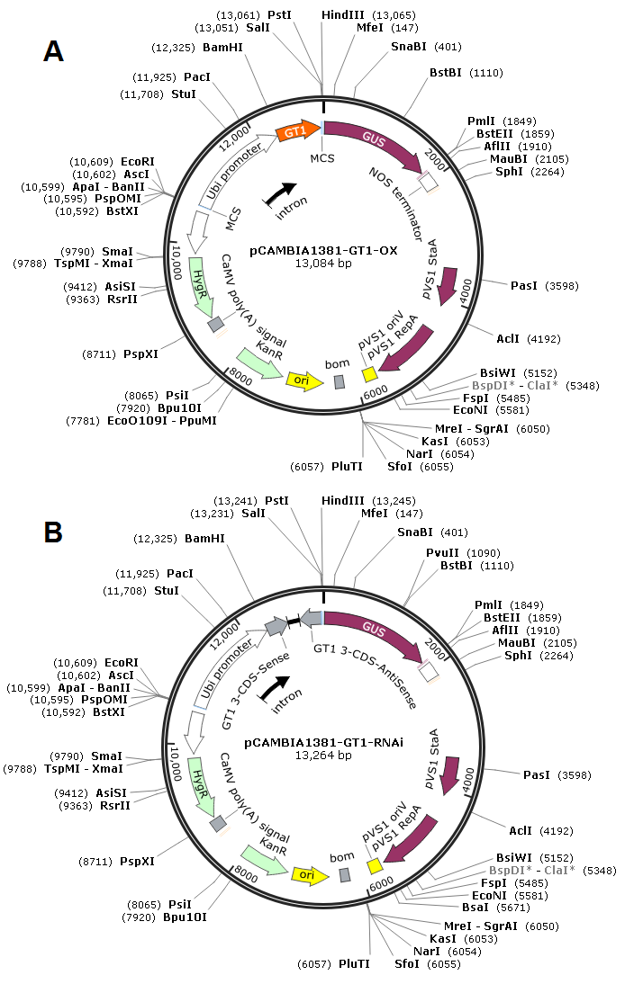
Vectors used for generating OsGT1 transgenic lines
The vectors used for generating OsGT1 overexpressors and RNA interference lines were visually illustrated using bioinformatics software Snapgene Version 2.3.2 (Fig. 3). pCAMBIA1381 is a binary vector used for Agrobacterium-mediated genetic transformation in plants. Numerous works [26-28] has successfully utilized this plasmid as backbone for constructing their design of interest. The transgenic plants were generated using pCAMBIA1381 and transformed using Agrobacterium tumefaciens methods [11].
The vector used in the generation of OX transgenic line was constructed by cloning the 720 bp open reading frame (ORF) of OsGT1 into the HindIII and SacI sites of the pCAMBIA1381 binary vector (Fig. 3 A). On the other hand, Ri transgenic plants were generated utilizing pCAMBIA1381 binary vector with the C-terminal 350 bp DNA fragment of OsGT1 both in the sense and antisense with a rice catalase intron cloned into the HindIII and SacI sites (Fig. 3 B).
Phenotype of OsGT1 transgenic lines
Phenotypic characteristics of OX and Ri were evaluated after maturation (160 DAP). The visual observation (Fig. 4) showed a that the plants overexpressing GT1 (OX) harbored fewer number of tillers along with a slight reduction in plant height in comparison to wild type and Ri counterparts. Despite Ri plants being notable higher, this height increase was not significant compared to the wild (NB) type. On the other hand, the height of OX plant was significantly lower than Ri and comparable to that of NB (Fig. 5 A). The reduction of plant height is a key objective in rice breeding, since shorter plant stature offers an ease of harvesting, tolerance against lodging stress, and an increased plant vigor upon rice grain maturity [30][31]. Moreover, shorter rice varieties could perform better under drought conditions [32][33]. The current results aligned with the finding of [25], who confirmed the plant height reduction in rice overexpressing OsGT1. The same study highlighted the role of GT1 gene along with HOX14 in regulating exsertion in panicle development and responding to endogenous gibberellic acid (GA) signaling. On the other hand, Ri lines from similar ZH11 background showed no difference to wild type in plant height [9]. Plant height significantly influences rice yield through two aspects [34]. The plant should uphold an upright position in tall plants to produce elongated panicles that can sustain a larger quantity of grains than a standard panicle can hold. The other aspect involves regulating plant height to reduce the danger of lodging caused by water stress or excessive grain packing in rice panicles. Rice panicles are reproductive structures composed of several individual flowers.

The panicle in rice is constituted by the formation of branch, spikelet, and floret meristems [35]. The length of a rice panicle typically correlates with the number of branches it can support. Consequently, in addition to the quantity of tillers, panicle length and branching are also significant determinants of rice yield [36]. In the current study, it was observed that neither overexpression nor down regulation of OsGT1 resulted in significant variations in panicle length or number of panicles per plant compared to the wild type (Fig. 5 B and C). Nevertheless, there was a slight reduction of panicle branches in GT1-OX in comparison to its wild type and Ri counterparts. Previous studies showed that elevated expression of Squamosa Promoter Binding Protein 14 (SPL14) gene in Nipponbare rice variety results in an increase in panicle branching [37][38] along with suppression of tiller number [38] enhancing yield in rice. This reflects a homeostatoic regulation of rice physiology to sustain a balance between panicle branching and “rice stem branching” through the generation of tillers [36].
The current observations confirmed prior research [11], indicating that GT1-OX plants exhibited a reduced number of tillers while GT1-Ri demonstrated an increased number of tillers relative to the wild-type (Fig. 5 D). No significant variations were seen in the percentage of productive tiller numbers between NB and GT1-OX (52%); however, GT1-Ri exhibited a markedly lower percentage of productive tiller numbers (41%) (Fig. 5 E). The quantity of productive tillers significantly influences photosynthate distribution, thereby ultimately determining the balance between green biomass and valuable grain yield, which is as the primary objective for farmers worldwide [6][39]. Subsequent research should focus on investigating factors that facilitate the conversion of non-productive tillers into productive ones.
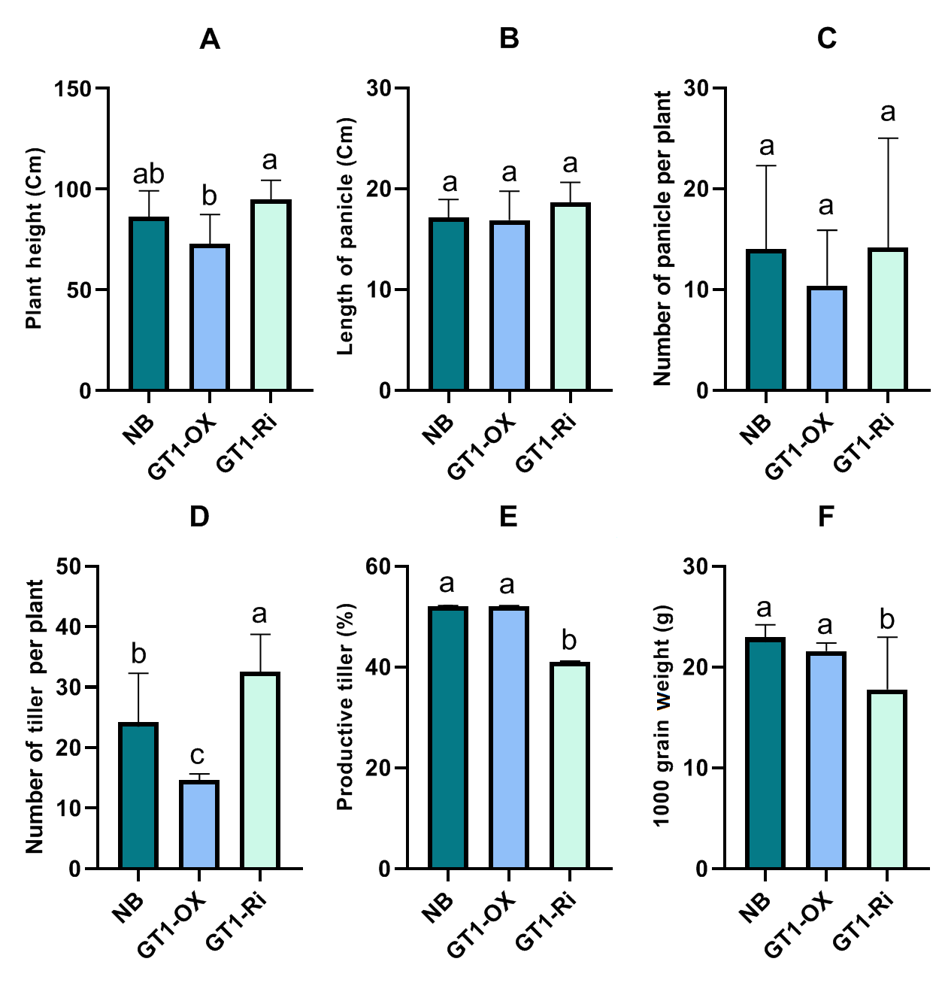
The observations revealed that the 1000 grain weight was significantly lower in Ri compared to that in NB and OX (Fig. 5 F). Moreover, there was a tendency for GT1-Ri to develop empty grain filling (data unpublished), hence influencing the final yield. A previous study [7] revealed that the use of rice harbouring tillering mutation can be beneficial under nutrient-deficiency conditions. The study confirmed that the rice mutant survived and maintained superior yield under stress conditions accompanied by phosphorus deficiency. However, the same research confirmed that while the number of grains increases with the increase in tiller number, achieving optimum grain filling remains significant challenges when utilizing rice variety with a higher number of tillers. Therefore, an integrative work and approach is fundamental to obtain high-yielding varieties to meet the global demands. Consequently, more research is necessary to assess the yield differences between NB, OX, and Ri in open field conditions.
Conclusions
This work provides an overview of the agronomical characterization of GT1-OX and GT1-Ri plants cultivated under greenhouse conditions. This study demonstrated that GT1 influences the number of tillers with the elevation of expression leading to a decrease in tiller number, while the reduction of expression leads to prolific growth of tillers. Further research is required to elaborate on the stability of the phenotype at a larger scale in tropical settings.
Acknowledgments
This research was supported by a grant from the PNBP (Grants No. 7394/PL17/KP/2024), Politeknik Negeri Jember. This work is a proceeding of a preliminary work presented at the 7th International Conference on Food and Agriculture 2024 (ICoFA 2024) (https://conference.polije.ac.id/icofa/2024/) hosted by Politeknik Negeri Jember, Indonesia.
References
- Khan MH, Dar ZA, Dar SA. Breeding Strategies for Improving Rice Yield—A Review. Agric. Sci. 2015;06:467–78. DOI
- Kumar A, Dixit S, Ram T, Yadaw RB, Mishra KK, Mandal NP. Breeding high-yielding drought-tolerant rice: Genetic variations and conventional and molecular approaches. J. Exp. Bot. 2014;65:6265–78. DOI
- Tanaka W, Yamauchi T, Tsuda K. Genetic basis controlling rice plant architecture and its modification for breeding. Breed Sci. 2023;73:22088. DOI
- Hussien A, Tavakol E, Horner DS, Muñoz-Amatriaín M, Muehlbauer GJ, Rossini L. Genetics of Tillering in Rice and Barley. Plant Genome. 2014;7. DOI
- Ordonio RL, Matsuoka M. New path towards a better rice architecture. Cell Res. 2017;27:1189–90. DOI
- Yan Y, Ding C, Zhang G, Hu J, Zhu L, Zeng D, Qian Q, Ren D. Genetic and environmental control of rice tillering. Crop J. 2023;11:1287–302. DOI
- Ishizaki T, Ueda Y, Takai T, Maruyama K, Tsujimoto Y. In-frame mutation in rice TEOSINTE BRANCHED1 (OsTB1) improves productivity under phosphorus deficiency. Plant Sci. 2023;330:111627. DOI
- Choi MS, Woo MO, Koh EB, Lee J, Ham TH, Seo HS, Koh HJ. Teosinte Branched 1 modulates tillering in rice plants. Plant Cell Rep. 2012;31:57-65. DOI
- Gao S, Fang J, Xu F, Wang W, Chu C. Rice HOX12 regulates panicle exsertion by directly modulating the expression of ELONGATED UPPERMOST INTERNODE1. Plant Cell. 2016;28:680–95. DOI
- Shao G, Lu Z, Xiong J, Wang B, Jing Y, Meng X, Liu G, Ma H, Liang Y, Chen F, Wang Y. Tiller bud formation regulators MOC1 and MOC3 cooperatively promote tiller bud outgrowth by activating FON1 expression in rice. Mol. Plant. 2019;12(8):1090-102. DOI
- Kumar V, Kim SH, Adnan MR, Heo J, Jeong JH, Priatama RA, Lee JJ, Kim CM, Je BI, Park SJ, Xuan YH. Tiller outgrowth in rice (Oryza sativa L.) is controlled by OsGT1, which acts downstream of FC1 in a PhyB-independent manner. J. Plant Biol. 2021;64(5):417-30. DOI
- Bin Rahman ANMR, Zhang J. Trends in rice research: 2030 and beyond. Food Energy Secur. 2023;12. DOI
- Agriculture F and FRCJ. Rice Annotation Project Database (RAP-DB). 2024. (Accessed August 5, 2024). Available from: Link
- Cold Spring Harbor Laboratory. Maize Gramene. 2024. (Accessed August 6, 2024). Available from: Link
- Cold Spring Harbor Laboratory. Sorghum Base. 2024. (Accessed August 5, 2024). Available from: Link
- European Molecular Biology’s Laboratory’s European Bioinformatics Institute. EnsemblPlants. 2024. (Accessed October 6, 2024). Available from: Link
- The Regents of University of California. Phytozome13. 2024
- European Bioinformatics Institute (EMBL-EBI), SIB Swiss Institute of Bioinformatics, Protein Information Resource (PIR). The Universal Protein Resource (UniProt). 2024.
- Whipple CJ, Kebrom TH, Weber AL, Yang F, Hall D, Meeley R, Schmidt R, Doebley J, Brutnell TP, Jackson DP. grassy tillers1 promotes apical dominance in maize and responds to shade signals in the grasses. Proc. Natl. Acad. Sci. 2011;108(33). DOI
- Kebrom TH, Spielmeyer W, Finnegan EJ. Grasses provide new insights into regulation of shoot branching. Trends. Plant Sci. 2013;18:41–8. DOI
- Dong Z, Alexander M, Chuck G. Understanding Grass Domestication through Maize Mutants. Trends Genet. 2019;35:118–28. DOI
- Gallagher JP, Man J, Chiaramida A, Rozza IK, Patterson EL, Powell MM, Schrager-Lavelle A, Multani DS, Meeley RB, Bartlett ME. GRASSY TILLERS1 (GT1) and SIX-ROWED SPIKE1 (VRS1) homologs share conserved roles in growth repression. Proc. Natl. Acad. Sci. 2023;120(51). DOI
- Sharif R, Raza A, Chen P, Li Y, El-Ballat EM, Rauf A, Hano C, El-Esawi MA. HD-ZIP gene family: potential roles in improving plant growth and regulating stress-responsive mechanisms in plants. Genes. 2021;12(8):1256. DOI
- Li Y, Yang Z, Zhang Y, Guo J, Liu L, Wang C, Wang B, Han G. The roles of HD-ZIP proteins in plant abiotic stress tolerance. Front. Plant Sci. 2022;13:1027071. DOI
- Shao J, Haider I, Xiong L, Zhu X, Hussain RM, Övernäs E, Meijer AH, Zhang G, Wang M, Bouwmeester HJ, Ouwerkerk PB. Functional analysis of the HD-Zip transcription factor genes Oshox12 and Oshox14 in rice. PLoS One. 2018;13(7). DOI
- Li J, Long Y, Qi GN, Li J, Xu ZJ, Wu WH, Wang Y. The Os-AKT1 channel is critical for K+ uptake in rice roots and is modulated by the rice CBL1-CIPK23 complex. Plant Cell. 2014;26(8):3387-402. DOI
- Ding C, Lin X, Zuo Y, Yu Z, Baerson SR, Pan Z, Zeng R, Song Y. Transcription factor OsbZIP49 controls tiller angle and plant architecture through the induction of indole‐3‐acetic acid‐amido synthetases in rice. Plant J. 2021;108(5):1346-64. DOI
- Hu L, Li H, Qin R, Xu R, Li J, Li L, Wei P, Yang J. Plant phosphomannose isomerase as a selectable marker for rice transformation. Sci. Rep. 2016;6(1):25921. DOI
- Snapgene. Plasmid Files – Snapgene. 2024. (Accessed August 6, 2024). Available from: Link
- Yuan L. Progress in super-hybrid rice breeding. Crop J. 2017;5:100–2. DOI
- Peng S, Khush GS, Virk P, Tang Q, Zou Y. Progress in ideotype breeding to increase rice yield potential. Field Crops Res. 2008;108:32–8. DOI
- Ahmadikhah A, Marufinia A. Effect of reduced plant height on drought tolerance in rice. 3 Biotech. 2016;6. DOI
- Rahmawati D, Dewi AK, Mufikasari VY, Wilujeng EDI, Adnan MochR. Agronomical Performances of Gajah Mungkur Mutant Rice Varieties Under Drought Stress. J. Trop. Life Sci. 2024;14:65–76. DOI
- Rahmawati D, Santika P, Fauzi DRR, Rosyadi Adnan dan M. Characterization of five local varieties of rice (Oryza sativa L.) in east java, Indonesia. In IOP Conference Series: Earth and Environmental Science. 2023;1168:012013. DOI
- Miao Y, Xun Q, Taji T, Tanaka K, Yasuno N, Ding C, Kyozuka J. ABERRANT PANICLE ORGANIZATION2 controls multiple steps in panicle formation through common direct-target genes. Plant Physiol. 2022;189(4):2210-26. DOI
- Liang W hong, Shang F, Lin Q ting, Lou C, Zhang J. Tillering and panicle branching genes in rice. Gene. 2014;537:1–5. DOI
- Miura K, Ikeda M, Matsubara A, Song XJ, Ito M, Asano K, Matsuoka M, Kitano H, Ashikari M. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 2010;42(6):545-9. DOI
- Luo L, Li W, Miura K, Ashikari M, Kyozuka J. Control of tiller growth of rice by osspl14 and strigolactones, which work in two independent pathways. Plant Cell Physiol. 2012;53:1793–801. DOI
- Wu G, Wilson LT, McClung AM. Contribution of rice tillers to dry matter accumulation and yield. Agron. J. 1998;90:317–23. DOI
Cite this article:
Adnan, M. R., Wilujeng, E. D. I., Aisyah, M. D. N., Alif, T., Galushasti, A. Preliminary agronomic characterization of japonica rice carrying a tiller number mutation under greenhouse conditions. DYSONA – Applied Science, 2025;6(2): 291-299. doi: 10.30493/das.2025.490594

