Musa Y Tula 1*; Onaiwu I Enabulele 2; Anthony E Ophori 2; Abumhere S Aziegbemhin 2; Osaretin Iyoha 3; Joel Filgona 4
1, Department of Biological Science Technology, Federal Polytechnic Mubi, Adamawa State, Nigeria
2, Department of Microbiology, Faculty of Life Sciences, University of Benin, Benin City, Edo State, Nigeria
3, Department of Medical Microbiology, School of Medicine, College of Medical Sciences, University of Benin, Benin City, Nigeria
4, Department of Microbiology, Adamawa State University Mubi, Adamawa State, Nigeria
E-mail:
birtyty@gmail.com
Received: 09/07/2022
Acceptance: 23/07/2022
Available Online: 24/07/2022
Published: 01/10/2022

Manuscript link
http://dx.doi.org/10.30493/DLS.2022.351097
Abstract
Water bodies constitute a medium through which antibiotic-resistant organisms and resistance genes are disseminated among the human and animal populations. In this research, Enterobacteriaceae species isolated from water sources in Adamawa-North Senatorial zone, Nigeria, were investigated with the focus on plasmid-mediated multi-drug resistance. The bacterial species were isolated from 256 water samples from well and river water sources by membrane filtration technique. The isolates were identified by cultural, identification kit, and molecular method. The Kirby-Bauer disc diffusion technique on Muller-Hinton agar was used to test their sensitivity to routinely used antibiotics. The multi-drug resistant Enterobacteriaceae were further screened for the presence of resistant plasmid by standard procedure. A total of 342 bacterial isolates were identified as members of Enterobacteriaceae, with Escherichia coli (24.6%) being the most predominant species. The sequences of the species documented in this study have been assigned accession numbers and have equally been added to the National Center for Biotechnology Information GenBank. Out of the 342 Enterobacteriaceae isolates, 293 (85.7%) were resistant to at least an antibiotic. Resistance to cotrimoxazole and ampicillin was the most common among the isolates, while resistance to imipenem was the lowest. The Multi-drug resistant (MDR) phenotype of all the isolates showed an overall prevalence of 32.5%. The isolates phenotyped as MDR were screened for plasmid profile. The results showed that 58 (65.2%) isolates harbored plasmid bands of various sizes. Three plasmid bands were detected on one isolate of P. mirabilis, two plasmid bands were detected in 10 isolates predominantly among E. coli, while 47 isolates harbored one plasmid band each. The potentially pathogenic bacteria carrying plasmid-mediated MDR phenotype in the water sources investigated in this study constitute a risk to the surrounding communities. Therefore, adequate intervention is needed to prevent the spread of this antibiotic-resistant bacteria to human communities.
Keywords: MDR, Drug-resistance, Enterobacteriaceae, Water, Adamawa
Introduction
Water is vital in every aspect of life [1] and is essential for healthy living [2]. Surface water is usually defined as any source of unprocessed freshwater which is often used for drinking, cooking, and other domestic purposes [3]. When water is contaminated by industrial waste, human waste, animal waste, garbage, untreated sewage, chemical effluents, and other pollutants, poor water quality becomes unavoidable.
Generally, drinking water is gradually becoming a limited resource due to the increase in population and per capita consumption. Additionally, water sources contamination caused by human activities is limiting drinking water resources even further [2][4]. When consumed, contaminated water may lead to several illnesses, including cholera, dysentery, diarrhea, typhoid, and other life-threatening chronic diseases. Therefore, the biological contamination of drinking water sources combined with fragile health systems pose a great threat to communities through disease outbreaks [5]. It is reported that 80% of the diseases worldwide occur due to contaminated water and water-borne pathogens [6][7].
Freshwater constitutes an encouraging living environment for several organisms, such as the Enterobacteriaceae group [8]. Bacterial isolates, particularly members of the Enterobacteriaceae, may enter aquatic bodies due to organic contamination from numerous sources, including but not limited to hospital effluents, wastewater treatment plants, discharge of livestock farms, and agricultural drainage. These activities introduce microorganisms in addition to doses of antibiotics into the water bodies [9][10]. As a result, water serves not only as a vehicle for the spread of antibiotic-resistant organisms among humans and animals, but also as a means of introducing resistance genes into natural bacterial ecosystems [8].
The presence of antimicrobial resistance genes in human and animal pathogens is of public and clinical significance, as it may lead to treatment failure, lengthy hospital stays, in addition to complex and lengthy treatment periods. These drug-resistant microbes may result in an imminent threat to life and economic losses [11][12]. It is estimated that infections with antibiotic-resistant bacteria are responsible for 700,000 annual deaths, and these numbers are projected to increase to 10,000,000 by the year 2050 [7][13].
Enterobacteriaceae is a group of Gram-negative rod-shaped bacteria, most of which are pathogenic and of clinical significance due to their disease-causing abilities. They demonstrate outstanding adaptive competence for acquiring and disseminating resistance genes [11] because they are amiable to changes. Mutational changes in bacterial isolates can be spread to susceptible cells by mobile genetic elements such as plasmids. Plasmids are extrachromosomal, self-directed DNA that may encode products that promote virulence or pathogenicity, spread drug resistance characters among a broad spectrum of bacteria, and serve as epidemiological markers [14].
The whole region of the Adamawa-North senatorial zone (comprising Mubi-North, Mubi-South, Maiha, Michika, and Madagali Local Government Areas) lacks pipe water supply, and groundwater from wells, private or public boreholes, in addition to rivers and streams are the primary water sources for cooking, drinking, and other domestic activities. The data regarding water-borne enteric as potential reservoirs for plasmid-mediated antimicrobial resistance in this region are scarce. Therefore, this study was carried out to determine the presence of enteric bacilli with multi-drug resistance (MDR) phenotype from water sources in the Adamawa-North senatorial zone of Adamawa State, Nigeria, and to ascertain whether this resistance trait is plasmid-borne.
Materials and Methods
Study Area
Adamawa North senatorial zone is commonly known as Mubi region. Mubi region comprises five local government areas, namely, Madagali, Michika, Mubi South, Mubi North, and Maiha, with a land size of 4,493.815k/m2 and a population of 682, 026. Mubi zone is located between latitudes 9o 30’ and 11o 00’N of the equator and between 13o 00’ and 14o 00E of the Greenwich meridian. The area has a tropical wet and dry climate. The dry season lasts for a minimum of six months (starting from November), while the wet season spans between May and October. The mean annual rainfall ranges from 700 mm to 1050 mm [15].
Water Sampling
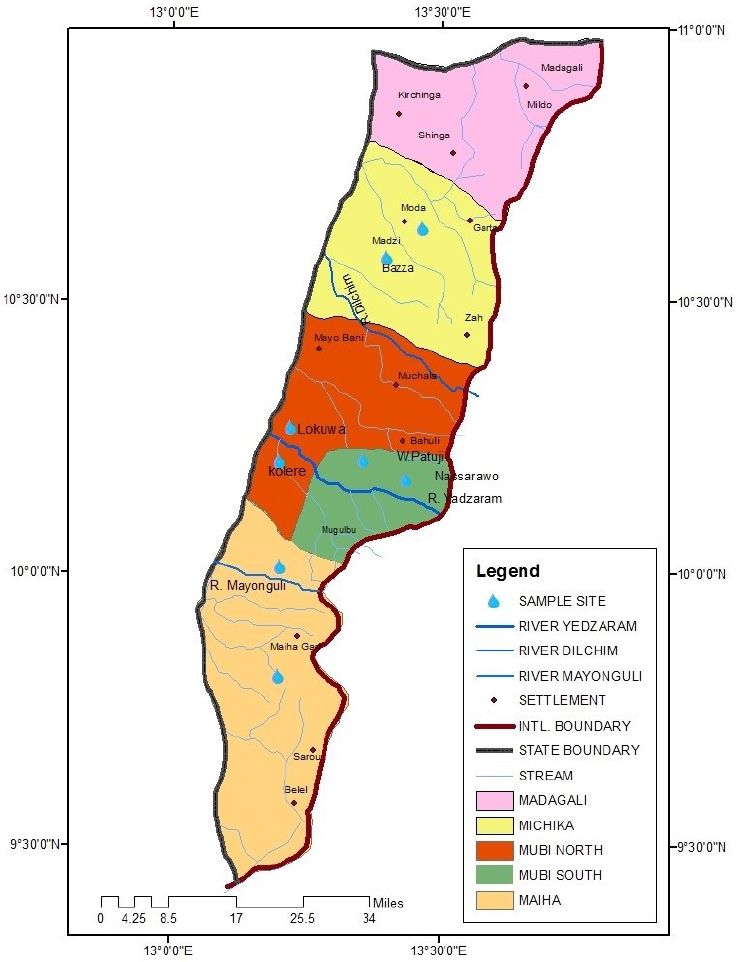
Water samples were collected from four local government areas (LGA) of Adamawa-North senatorial zone. Well water samples were taken from 8 locations, two from each LGA: Lokuwa and Kolere from Mubi-North, Wuro-patuji and Nassarawo from Mubi-South, Michika and Bazza from Michika LGA, Maiha and Pakka from Maiha LGA. River water samples were taken from four rivers, one from each local government area: river Yadzaram (Mubi-North and South), river Dilchim (Michika LGA), and river Mayonguli (Maiha LGA) (Fig. 1). Samples were taken from June to October 2019 (rainy season), and January-May, 2020 (dry season). 100 ml sterilized screw cap sampling bottles (Bijou bottles) were used for sample collection. Overall, 32 water samples were collected from each location (a total of 256 samples).
Isolation of Enterobacteriaceae species
Enterobacteriaceae species were isolated by the membrane filtration technique. For each sample, a sterile mixed cellulose ester (MCE) membrane filter (Merck, Bangalore) with a diameter of 47 mm and a pore size of 0.45 μm was placed on the porous base of the filtration unit using sterile forceps. 50 mL of the water sample were dispensed into the funnel of the filtration unit, and filtration was achieved by a manual suction pump. At the end of the filtration, the funnel was dismantled; a sterile forceps was used to pick the filter onto the surface of MacConkey agar (MCA) and replicated on Salmonella-Shigella agar (SSA), Eosin methylene blue (EMB) agar, and Deoxycholate citrate agar (DCA). The plates were incubated at 35 – 37 oC for 18 – 24 h. Discrete bacterial colonies were Gram-stained, re-cultured, and stored in nutrient agar slant for identifications and further use.
Biochemical identification of bacterial isolates
After Gram-staining, the presumptive identification of Enterobacteriaceae was carried out using standard biochemical tests, and the isolates were confirmed using Microgen Gram negative-A (GN-A) ID kits.
Molecular identification of bacterial isolates
After genomic DNA extraction, the 16S rRNA gene was amplified and purified. The amplified fragments were cleaned using ethanol, and their reliability was proven on 1% agarose gel [16]. The sequencing of the amplified fragments was achieved on a Genetic Analyzer 3130 x l sequencer (Applied Biosystems) at Inqaba Diagnostic, South Africa. The isolates’ identity was confirmed by subjecting the sequences to analysis with the Basic Local Alignment Search Tool (BLAST). Available at https://www.ncbi.nlm.nih.gov/blast.
Antibiotic susceptibility of bacterial isolates
The susceptibility profile of the isolates to frequently used antibiotics was determined on Mueller-Hinton agar (Oxoid, UK) in vitro by the disc diffusion method. The following antibiotic discs were used: pefloxacin (10 μg), gentamicin (10 μg), ofloxacin (10 μg), imipenem (10 μg), ceftriaxone (30 μg), ceftazidime (30 μg), streptomycin (30μg), ciprofloxacin (10 μg), augmentin (30 μg), ampicillin (30 μg), nalidixic acid (30 μg), and cotrimoxazole (30 μg). Well-separated colonies of the bacterial isolates on nutrient agar were picked by a sterile wire loop and mashed in sterile normal saline to match the 0.5 McFarland turbidity standard. Afterward, the suspension was smeared uniformly on the surface of the prepared Mueller-Hinton agar plate. Each antibiotic disc was placed on the surface of the inoculated plate using sterile forceps, and the plates were incubated for 18-24hrs at 37oC. The inhibition zone diameter was measured in millimeters and was interpreted based on the diameter interpretative standard breakpoints [17]. MDR phenotypes were determined when an isolate was resistant to at least three different classes of antibiotics.
Plasmid DNA isolation and agarose gel electrophoresis
For plasmid DNA extraction from the selected bacterial isolates, QIAGEN Plasmid Purification mini kit was employed [18] on the principles of the alkaline lysis method [19], and the reliability of the extracted plasmids was proven on 1% agarose gel.
Statistical analysis
Data generated were presented in percentages. The antibiotic resistance means were compared using the least significance difference (LSD) test at P<0.05. All data were analyzed using the Statistical Package for Social Sciences version 17.
Results
Enterobacteriaceae isolates distribution
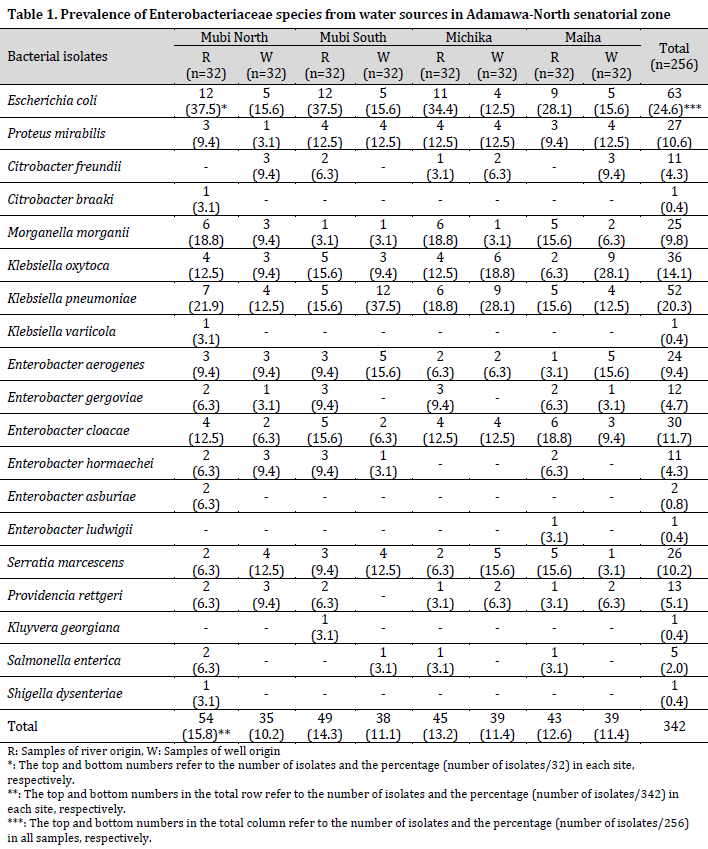
From the 256 water samples analyzed for bacteria growth, 342 isolates were identified as members of the Enterobacteriaceae family (189 from the rainy season and 153 from the dry season), belonging to 11 genera and 19 species. The species of the Enterobacteriaceae family include Escherichia coli, Proteus mirabilis, Citrobacter freundii, Citrobacter braaki, Morganella morgani, Klebsiella oxytoca, Klebsiella pneumoniae, Klebsiella variicola, Enterobacter aerogenes, Enterobacter gergoviae, Enterobacter cloacae, Enterobacter hormaechei, Enterobacter asburiae, Enterobacter ludwigii, Serratia marcescens, Providencia rettgeri, Kluyvera georgiana, Salmonella enterica, and Shigella dysenteriae (Table 1).
Escherichia coli was the most predominant bacterial species and was isolated from 24.6% of the samples, closely followed by K. pneumoniae (20.3%) and K. oxytoca (14.1%). At the same time, the least prevalent species were C. braaki, K. variicola, K. georgiana, E. ludwigii, and Shigella dysenteriae, with one isolate and a prevalence of 0.4% each. The number of bacterial isolates recovered from river water sources was higher than those from well water sources but with no significant difference (P=0.081). However, the majority of the bacterial species were recovered from the river Yadzaram that transverse between Mubi-South and North LGA. Also, a higher number of bacterial isolates was recorded in Mubi-North (26%) and Mubi-South (25.4%), while Maiha recorded the least number of isolates (24%).
The results showed that Citrobacter braaki, Klebsiella variicola, Enterobacter aerogenes, Serratia marcescens, Providencia rettgeri, and Kluyvera georgiana were only recovered in the rainy season. In contrast, Citrobacter freundii, S. enterica, Shigella dysenteriae, Enterobacter gergoviae, Enterobacter hormaechei, Enterobacter asburiae, and Enterobacter ludwigii were only recovered in the dry season. Other species of bacteria were recovered variably in both seasons. The sequences of the Enterobacteriaceae documented in this study have been assigned accession numbers and added to the National Center for Biotechnology Information (NCBI) GenBank (Supplementary Table 1).
Enterobacteriaceae drug-resistance
Drug resistance tests showed that out of the 342 Enterobacteriaceae isolates, 49 (14.3%) were sensitive to all the tested antibiotics, while 293 (85.7%) bacteria species were resistant to at least an antibiotic. Furthermore, a variable resistance pattern to all the tested antibiotics was observed among the resistant isolates. Escherichia coli, P. mirabilis, E. gergoviae, and E. asburiae were mostly resistant to ampicillin, closely followed by cotrimoxazole. At the same time, K. oxytoca, K. pneumoniae, M. morgani, P. rettgeri, S. marcescens, E. aerogenes, and E. cloacae were predominantly resistant to cotrimoxazole, closely followed by ampicillin. Citrobacter braaki was resistant only to cotrimoxazole, ceftriaxone, and ceftazidime, while K. georgiana was only resistant to ceftriaxone and ceftazidime. Enterobacter ludwigii and K. variicola were resistant to all the tested antibiotics, while Shigella dysenteriae was resistant to all antibiotics except pefloxacin (Table 2).
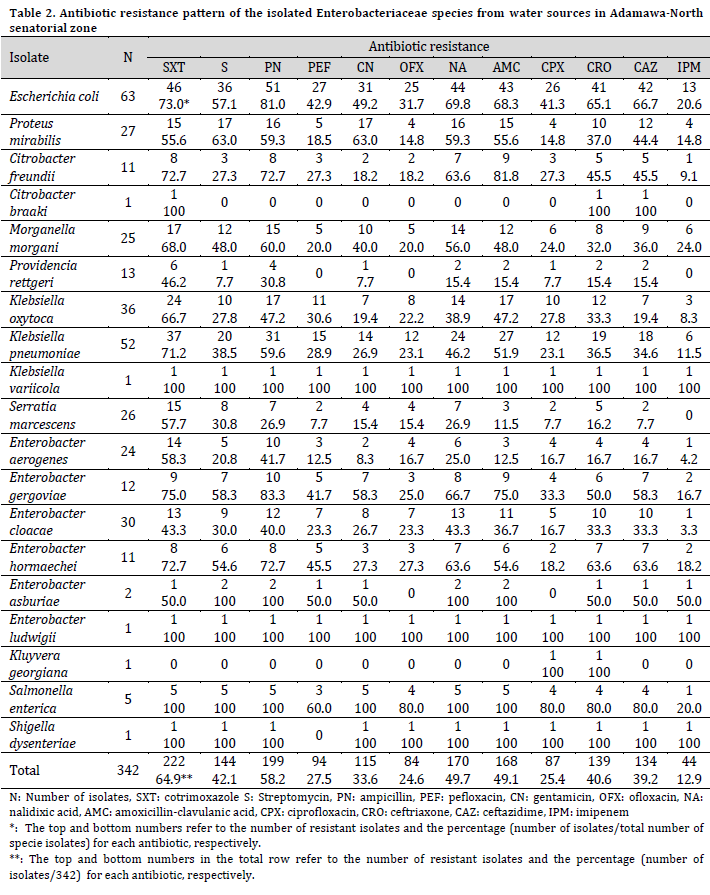
The resistance to cotrimoxazole was the highest among the studied isolates, with no statistical difference from those of streptomycin, ampicillin, nalidixic acid, amoxicillin-clavulanic acid, ceftriaxone, and ceftazidime (P=0.079). On the other hand, resistance to imipenem was the lowest but with no statistical difference from those of pefloxacin, gentamicin, ofloxacin, and ciprofloxacin (P=0.102).
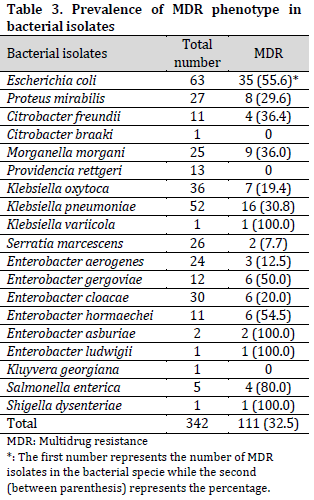
The Multi-drug resistant (MDR) phenotype of all the bacterial isolates showed an overall prevalence of 32.5%. From these, all the species of K. variicola, E. asburiae, E. ludwigii, and Shigella dysenteriae exhibited the MDR phenotype. Also, the isolates of S. enterica (80.0%), E. coli (55.6%), E. hormaechei (54.5%), and E. gergoviae (50.0%), among others, exhibited MDR phenotype in different proportions (Table 3). The results further showed that none of the isolates of C. braaki, P. rettgeri, and K. georgiana exhibited MDR phenotype.
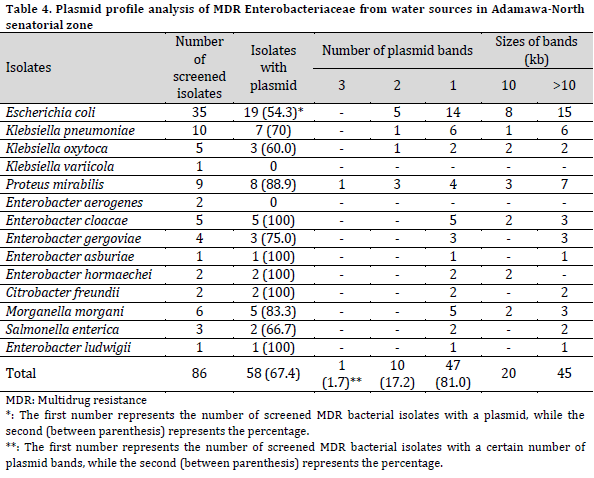
Plasmid profile in MDR isolates
The isolates phenotypes as MDR Enterobacteriaceae were screened for plasmid profile. Among these isolates, 58 (65.2%) harbored plasmid bands of various sizes. All the isolates of E. ludwigii, C. freundii, E. hormaechei, and E. asburiae harbored plasmid bands. Also, the isolates of P. mirabilis (88.9%), M. morgani (83.3%), E. gergoviae (75.0%), K. pneumoniae (70.0%), S. enterica (66.7%), K. oxytoca (60.0%), and E. coli (54.3%) harbored plasmid. Furthermore, no plasmid bands were detected in either K. variicola or E. aerogenes. The Enterobacteriaceae were categorized into three groups based on the number of plasmid bands (3, 2, and 1 plasmid bands) in each isolate. Three plasmid bands were detected in only one isolate of P. mirabilis. Two plasmid bands were detected in 10 isolates with the highest occurrence in E. coli, closely followed by P. mirabilis, while 47 isolates harbored one plasmid band each.
The molecular weights of the plasmids were ≥10kb. The molecular weight of detected plasmid bands was >10kb in 45 isolates, while 20 MDR isolates carried a 10kb plasmid (Table 4).
Discussion
Among theEnterobacteriaceae, Escherichia coli was the most prevalent bacterial species encountered in this study. This predominance implies pollution of the water samples understudy with fecal matter. This finding is in accordance with another study in Jimeta-Yola, Adamawa State, where various bacterial species were recovered from water samples, with E. coli being the most prevalent [20]. Similarly, multiple works reported the preponderance of E. coli in water sources in Bosso town, Minna [21], Benue State [22], Kano State [23], Ebonyi State [24], Addis-Ababa, Ethiopia [25], Pakistan [26], Eastern Cape region of South-Africa [27]. On the other hand, several studies reported the predominance of Klebsiella species over other bacterial isolates in their study area [28][29] which was contrary to the finding of this study. Also, a high prevalence of Enterobacter spp. from a water source was reported in Ijebu-Ode, Southwestern Nigeria [30]. Furthermore, Enterobacter and Klebsiella spp., were reported in communal water sources in Ogun State, Nigeria [11], while the preponderance of Proteus and Klebsiella spp. in treated and untreated water samples, respectively, were reported in South-West Nigeria [31]. Similarly, a study in Ahiazu Mbaise, Eastern Nigeria, reported the preponderance of Salmonella and Shigella spp. over other Enterobacteriaceae in various water sources [32].
During the rainy season, a higher number of bacterial isolates, particularly E. coli, were recovered than in the dry season. This finding corroborates previously reported observations of an increase in bacterial number in water bodies during rainfall [29][33][34]. This observation could be attributed to the high water influx in both water sources during the rainy season, which carries organic and inorganic pollutants and accumulates them in the water sources [35][36].
Typical enteric pathogens isolated from water sources in this study are Shigella dysenteriae and Salmonella enterica. These pathogens are aetiologic agents of shigellosis, typhoid/paratyphoid, or gastroenteritis. Other bacterial species may be pathogenic in immunocompromised individuals and children or infants. Detecting a large number of varying enteric pathogens in the river and well water sources in the study area may suggest the continual spread of enteric bacteria with the potential of causing life-threatening diseases in the community, particularly among children, the elderly, and immuno-compromised individuals.
Most of the isolated enteric pathogens were highly resistant to ampicillin and/or cotrimoxazole. High antibiotic resistance was similar to previous studies in Abakaliki [24] and Ghana [37]. High resistance to ampicillin [38- 40] and cotrimoxazole [41] by enteric pathogens in water sources were also reported previously. Low resistance to imipenem among Enterobacteriaceae in this study is reasonably expected since imipenem was not used in the study area at the time of this research. However, even these low resistance rates among the studied isolates can also be alarming and might suggest the occurrence of cross-resistance. Cross-resistance occurs when an organism is resistant to an antibiotic to which not earlier exposed, probably due to the acquisition of resistance markers to other antibiotics [42][43]. The high prevalence of resistance to commonly used antibiotics such as ampicillin, cotrimoxazole, streptomycin, nalidixic acid, amoxicillin-clavulanic acid, ceftriaxone, and ceftazidime in this study constitute a public health risk because these antibiotics may not be effective in treating individuals who utilized the examined water sources for drinking or other domestic purposes.
In this regard, [44] documented the high prevalence of multi-drug resistant Gram-negative bacterial infections in Sokoto, Northwest Nigeria, predominantly caused by E. coli and K. pneumoniae. The majority of the isolates were MDR and showed extended spectrum β-lactamase (ESβL) and carbapenemase activities. By comparing [44] results and those from the current study, a strong positive correlation (r = 0.907) can be found in the MDR nature for E. coli from patient isolates reported by [44] and water isolates in the current study (Fig. 2). This correlation suggests high similarities in MDR profiles between contagious bacterial species in the country. Therefore, immediate implementation of infection control procedures and antibiotic stewardship in hospitals is needed in order to prevent the spread of antibiotic-resistant bacteria inside and outside healthcare facilities.
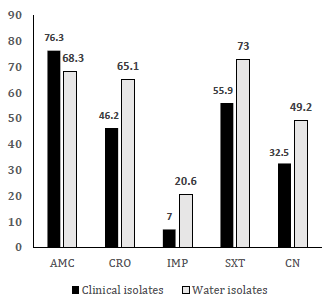
Figure 2. MDR correlation between clinical [44] and water sources isolates of E. coli in Nigeria
The agarose gel electrophoresis results showed the presence of plasmid DNA bands in the inspected MDR Enterobacteriaceae species in this study. Similarly, a previous study revealed that plasmids are frequently found to unpredictable extents in bacterial isolates from water sources, with various frequencies [45]. Other studies reported the detection of plasmids in isolates from wastewater [46][47]. Most MDR isolates harbored one plasmid, while others had multicopy plasmid bands. This finding is similar to the reports of previous studies [14][46][48]. The detected plasmids in the MDR isolates of this study are not only evidence of resistance genes but also suggest that the isolates can obtain other determinants as a survival strategy and may be able to transfer these traits to other susceptible bacterial species [47][49].
The plasmid profile of the MDR Enterobacteriaceae had shown the presence of high molecular weight (≥10kb) plasmid bands. Plasmids that mediated MDR phenotype in bacterial species were reported to be of high molecular weight [14]. Thus, it can be concluded that most MDR bacterial species harboring high molecular weight plasmids may carry other resistant determinants in addition to other virulent genes. Therefore, the study of plasmid profile is of high significance, as it provides means through which bacteria-carrying plasmids are associated with specific phenotypic attributes, either virulent or drug-resistant determinants.
The whole of the Adamawa-North senatorial zone (comprising Mubi-North, Mubi-South, Maiha, Michika, and Madagali Local Government Areas) lacked pipe-borne water supply. Therefore, the primary water sources for cooking, drinking, and other domestic activities are groundwater from wells, private or public boreholes, and rivers/streams. As such, the presence of potentially pathogenic bacteria carrying plasmid-mediated MDR phenotype in the water sources investigated may imply that antibiotic-resistant traits can easily and rapidly spread from resistant to susceptible bacterial species in the ecosystem. Consumption of such water without treatment may also constitute a potential risk of community-acquired infections, which may lead to an outbreak of disease and the spread of antibiotic-resistant bacteria in human and animal populations.
Conclusion
This study detected the species of Enterobacteriaceae in water sources by phenotypic and molecular methods and underscored the public health significance of plasmid-borne MDR Enterobacteriaceae in the water sources, especially if consumed untreated.
Therefore, Immediate interventions in infection control and antibiotic stewardship in hospitals are needed to prevent the spread of antibiotic-resistant bacteria inside and outside healthcare facilities. Furthermore, adequate water treatment is essential to prevent a possible looming outbreak of antibiotic-resistant bacterial-borne diseases in human and animal populations.
References
| 1 | Mishra M, Arukha AP, Patel AK, Behera N, Mohanta TK, Dhananjay Y. Multi-Drug Resistant Coliform: Water Sanitary Standards and Health Hazards. Front. Microbiol. 2018;9. DOI |
| 2 | Hassan-Rashid MAU, Manzoor MM, Mukhtar S. Urbanization and its effects on water resources: An exploratory analysis. Asian J. Water Environ. Pollut. 2018;15:67–74. DOI |
| 3 | Vaz-Moreira I, Egas C, Nunes OC, Manaia CM. Bacterial diversity from the source to the tap: A comparative study based on 16S rRNA gene-DGGE and culture-dependent methods. FEMS Microbiol. Ecol. 2013;83:361–74. DOI |
| 4 | Levantesi C, Bonadonna L, Briancesco R, Grohmann E, Toze S, Tandoi V. Salmonella in surface and drinking water: Occurrence and water-mediated transmission. Food Res. Int. 2012;45:487-602. DOI |
| 5 | World Health Organisation (2018). Fact Sheet: Drinking Water. Geneva: World Health Organization |
| 6 | Haseena M, Malik MF, Javed A, Arshad S, Asif N, Zulfiqar S, Hanif J. Water pollution and human health. Environ. Risk Assess. Remed. 2017;1(3):16-9. |
| 7 | Singh AK, Das S, Kumar S, Gajamer VR, Najar IN, Lepcha YD, Tiwari HK, Singh S. Distribution of Antibiotic-Resistant Enterobacteriaceae Pathogens in Potable Spring Water of Eastern Indian Himalayas: Emphasis on Virulence Gene and Antibiotic Resistance Genes in Escherichia coli. Front Microbiol. 2020;11:581072. DOI |
| 8 | Djenadi K. Antibiotic Resistance Bacteria from Rivers Water in Algeria. J Bacteriol Parasitol. 2017;8:319. DOI |
| 9 | Aydin S, Aydin ME, Ulvi A, Kilic H. Antibiotics in hospital effluents: occurrence, contribution to urban wastewater, removal in a wastewater treatment plant, and environmental risk assessment. Environ. Sci. Pollut. Res. Int. 2019;26(1):544–58. DOI |
| 10 | Cohen R, Paikin S, Rokney, A, Blum M.R. and Astrahan P. Multidrug-resistant Enterobacteriaceae in coastal water: an emerging threat. Antimicrob. Resist. Infect. Control. 2020;9:169. DOI |
| 11 | Odumosu BT, Akintimehin AR. Occurrence of Extended-Spectrum Beta-Lactamase producing Enterobacteriaceae isolates in communal water sources in Ogun State, Nigeria. African J. Clin. Exp. Microbiol. 2015;16(1):28–32. DOI |
| 12 | Bueno I, Williams-Nguyen J, Hwang H, Sargeant JM, Nault AJ, Singer RS. Systematic Review: Impact of point sources on antibiotic-resistant bacteria in the natural environment. Zoonoses Public Health. 2018;65:162–84. DOI |
| 13 | Morris S, Cerceo E. Trends, epidemiology, and management of multi-drug resistant gram-negative bacterial infections in the hospitalized setting. Antibiotics. 2020;9:1–20. DOI |
| 14 | Lina TT, Rahman SR, Gomes DJ. Multiple-Antibiotic resistance Mediated by Plasmids and Integrons in Uropathogenic Escherichia coli and Klebsiella pneumoniae. BangladeshJ Microbiol. 2007;24(1):19-23. DOI |
| 15 | Adebayo AA, editor. Mubi region: a geographical synthesis. Paraclete; 2004:17-25 |
| 16 | Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ. Critical Evaluation of Two Primers Commonly Used for Amplification of Bacterial 16S rRNA Genes. Appl. Environ. Microbiol. 2008;74:2461-70. DOI |
| 17 | Wayne A. Clinical and Laboratory Standards Institute; CLSI. 2011. Performance standards for antimicrobial susceptibility testing. 20th Informational Supplement. CLSI document. 2017. |
| 18 | Midi QP, Maxi M, Kits G. QIAGEN® Plasmid Purification Handbook. 2000. |
| 19 | Birnboim HC, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7(6):1513-23. DOI |
| 20 | Bala JD. Occurrence of faecal coliform in water sources in Jimeta-Yola. J Environ. Sci.2006;10(2):64-66. |
| 21 | Oyedum UM, Adabara NU, Oloruntimilehin BJ. A survey of bacteriological quality of well water from various locations in Bosso town, Northcentral, Nigeria. FUTA J. Res. Sci. 2016;12(2):280–6 |
| 22 | Ichor TEU, Umeh EU, Duru EE. Microbial Contamination of Surface Water Sources in Rural Areas of Guma Local Government Area of Benue State, Nigeria. J. Med. Sci. Pub. Health. 2014;2(2):43-51 |
| 23 | Taura DW, Hassan A. Bacteriological examination of households drinking water in some local Government areas of Kano State, Nigeria. Intl. Res. J. Pharm. Pharmacol. 2013; 3(6):91-6 |
| 24 | Okeke OCO, Romanus II, Anthony NI, Nnabuife A, Obehi E, Chijindu OS. Antimicrobial resistance pattern of coliform bacteria isolated from sachet and borehole waters sold in Abakaliki metropolis of Ebonyi State, Nigeria. Int. J. Innov. Sci. Res. 2015;16(2):526-32 |
| 25 | Tesfaye H, Alemayehu H, Desta AF, Eguale T. Antimicrobial susceptibility profile of selected Enterobacteriaceae in wastewater samples from health facilities, abattoirs, downstream rivers and a WWTP in Addis Ababa, Ethiopia. Antimicrob. Resist. Infect. Control. 2019;8:134. DOI |
| 26 | Humayun E, Bibi A, Rehman AU, Ahmad S, Shujaat N. Isolation and Identification of Coliform Bacteria from Drinking Water Sources of Hazara Division, Pakistan. IOSR J Pharm. 2015;5(4):36-40 |
| 27 | Muringani BN, Obi CL, Apalata T, Vasaikar SD. Prevalence of potential enteric pathogens in treated and untreated water sources around Eastern Cape region. J. Bacteriol Mycol Open Access, 2015;1(1):14‒9. DOI |
| 28 | Ugochukwu S, Giwa FJ, Giwa A. (2015). Bacteriological evaluation of sampled sachet water sold in Samaru-Zaria, Kaduna-State, Nigeria. Nigerian J. Basic Clin. Sci. 2015; 12:6-12. DOI |
| 29 | Odonkor ST, Addo KK. Prevalence of Multidrug-Resistant Escherichia coli isolated from Drinking Water Sources.Intl. J. Microbiol. 2018;720401. DOI |
| 30 | Bello OO, Osho A, Bello TK. Microbial quality and antibiotic susceptibility profiles of bacterial isolates from borehole water used by some schools in Ijebu-Ode, Southwestern Nigeria. Scholars Acad. J. Biosci. 2013;1(1):4-13 |
| 31 | Adesoji AT, Ogunjobi AA. Detection of Extended Spectrum Beta-Lactamases Resistance Genes among Bacteria Isolated from Selected Drinking Water Distribution Channels in Southwestern Nigeria. BioMed Res. Int. 2016:7149295. DOI |
| 32 | Esomonu OC, Abanobi OC, Ihejirika CE. Enteric pathogens and diarrhoea disease potentials of water sources in Ahiazu Mbaise, Eastern Nigeria. J. Public Health Epidemiol.2012;4(2):39-43. DOI |
| 33 | Parker JK, McIntyre D, Noble RT. Characterizing faecal contamination in stormwater runoff in coastal North Carolina, U.S.A. Water Res. 2010;44(14):4186–94. DOI |
| 34 | Sidhu JPS, Ahmed W, Hodgers L, Toze S. Occurrence of Virulence Genes Associated with Diarrheagenic Pathotypes in Escherichia coli Isolates from Surface Water. Appl. Environ. Microbiol. 2013;79(1):328-35. DOI |
| 35 | Kisteman T, Claben T, Roch C, Dangeradorf F, Fisheder R, Gebel J, et al. Microbial load of Drinking water Reservoir Tributaries during Extreme rainfall and runoff. Appl .Environ. Microbiol. 2002;68:2188-97. |
| 36 | Garba I, Tijjani MB, Aliyu MS, Yakubu SE, Wada-Kura A, Olonitola OS. Prevalence of Escherichia coli in some public water sources in Gusau Municipal, North-Western Nigeria. Bayero J. Pure Appl. Sci. 2009;2(2):134-7 |
| 37 | Tagoe DNA, Nyarko H, Arthur SA, Birikorang E. A study of antibiotic susceptibility pattern of bacterial isolates in sachet drinking water sold in Cape Town Coast Metropolis of Ghana. Res. J. Microbiol. 2011;6(2):153-8. DOI |
| 38 | Nimri LF, Azaizeh BA. First report of multidrug-resistant ESBL producing urinary Escherichia coli in Jordan. British Microbiol. Res. J.2012;2(2):71-81. DOI |
| 39 | Ogunleye AG, Omoya FO, Ayantola KJ. Bacterial antibiogram and physicochemical parameters of well water in Iworoko-Ekiti, Nigeria. J. Appl. Life Sci. Intl. 2016;4(4):1-10. DOI |
| 40 | Ogu GI, Madar IH, Olueh AA, Tayubi IA. Antibiotic Susceptibility Profile of Bacteria Isolated from Drinking Water Sources in Amai Kingdom, Delta State, Nigeria. Annual Res. Rev. Biol. 2017;14(1):1-9. DOI |
| 41 | Sharma BC, Rai B. Incidence of multi-drug resistance in Escherichia coli strains isolated from three lakes of tourist attraction (Mirik lake, Jorepokhari lake and Nakhapani lake) of Darjeeling hills, India. Indian J. Fundam. Appl. Life Sci. 2012;2:108–14. |
| 42 | Levy SB, Marshall B. Antibacterial resistance worldwide: Causes, challenges and responses. Nat. Med. 2004; 10(12):122-129. DOI |
| 43 | Tripathi KD. Essentials of medical pharmacology. JP Medical Ltd. 2013. |
| 44 | Olowo-okere A, Ibrahim YKE, Nabti LZ, Olayinka BO. High prevalence of multidrug-resistant Gram-negative bacterial infections in Northwest Nigeria. Germs. 2020;10(4):310-21. DOI |
| 45 | Banu H, Prasad KP. Role of plasmids in Microbiology. J. Aquac. Res. Dev. 2017; 8(1):1-8. DOI |
| 46 | Pignato S, Coniglio MA, Faro G, Weill FX, Giammanco G. Plasmid-mediated multiple antibiotic resistance of Escherichia coli in crude and treated wastewater used in agriculture. J. Water Health. 2009;7(2):251-8. DOI |
| 47 | Li Q, Chang W, Zhang H, Hu D, Wang X. The Role of Plasmids in the Multiple Antibiotic Resistance Transfer in ESBLs-Producing Escherichia coli Isolated from Wastewater Treatment Plants. Front. Microbiol. 2019;10:633. DOI |
| 48 | Wang H, Mao X, Ding H, Zou Q, Peng X. Epidemiological survey on Escherichia coli O157 in Chongqing and Three-Gorge Reservoir Areas of China. Vet. Res. Commun. 2008;32(6):449-61. DOI |
| 49 | Mokracka J, Koczura R, Kaznowski A. Multi resistant Enterobacteriaceae with class 1 and class 2 integrons in a municipal wastewater treatment plant. Water Res. 2012; 46:3353. DOI |
Cite this article:
Tula, M., Enabulele, O., Ophori, A., Aziegbemhin, A., Iyoha, O., Filgona, J. Phenotypic and molecular detection of multi-drug resistant Enterobacteriaceae species from water sources in Adamawa-North senatorial zone, Nigeria. DYSONA – Life Science, 2022;3(2): 57-68. doi: 10.30493/dls.2022.351097
